Copper and Iron Metabolism
The Vulpe Lab is also involved in collaborations to study the genetic determinants of iron deficiency in humans.The Vulpe group investigates hephaestin, a protein that serves as a key intersection between copper and iron metabolism in mammals by taking advantage of the remarkable conservation of fundamental cellular mechanisms across species through the use of functional profiling methods, such as CRISPR technology, in model organisms to gain insight into copper and iron metabolism.
Hephaestin
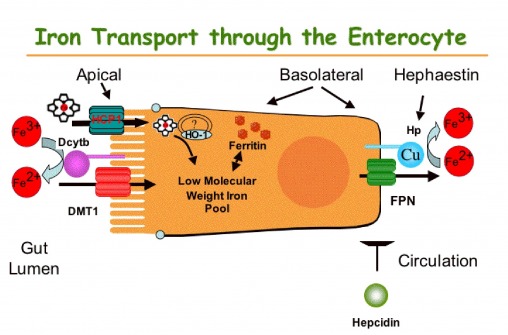
Iron absorption by the intestine controls iron homeostasis in mammals. Intestinal iron absorption is remarkably balanced to provide adequate iron to meet the body’s iron needs while preventing toxic excess. Because humans have an extremely limited capacity to excrete iron, regulation of the body’s iron levels is achieved by modulating uptake.
We identified the gene that is mutant in mice with sex linked anemia. Hephaestin (Hp) was predicted to be a membrane-bound copper containing ferrroxidase Because sla mice have defective intestinal iron efflux, we hypothesized that the proposed ferroxidase activity provided by hephaestin may facilitate efficient iron export. The group has for the last several years undertaken to systematically test this hypothesis to better understand hephaestin’s central role in intestinal iron metabolism Overall, this work has confirmed our hypotheses that hephaestin is a membrane bound multi-copper ferroxidase required for export of iron from the intestinal enterocyte into the cirulation and is providing the foundation and intriguing leads for further understanding the role of hephaestin in intestinal iron transport. Iron requirements regulate the amount of iron entering the body via intestinal absorption. In a continued collaboration with Dr. G Anderson (QIMR, Brisbane, Australia) and a new collaboration with Dr. McKie (King’s College, London, U.K.), we have begun to provide a molecular understanding of these regulatory processes. In a critical step to dissect this complex regulatory system, we identified the molecular targets of systemic regulation in a mature intestinal absorptive cells as the basolateral components, Hephaestin and Ireg1. We proposed a model of intestinal iron metabolism in which two layers of regulation allows for a modulated response to changes in iron status to ensure adequate iron uptake and minimize potential toxicity.
Iron is an essential metal, involved in many processes of the electron transport with transport of oxygen being the most important. Lack of known regulated mechanisms of iron excretion and iron’s toxicity, when in excess underline the importance of a fine balance between iron absorption, utilization and loss. The disturbances in this process can result in iron overload or iron deficiency. Numerous studies on human, mice and other species indicate a significant genetic contribution to iron homeostasis involving multiple loci.
Human Studies
We here focus on iron deficiency, which is the most common disease in the world with estimated 4-5 billion affected persons. Consequences of iron deficiency run from altered immune function, poor work performances and fatigue to anemia in severe cases. There is a strong environmental influence in the development of iron deficiency, with insufficient dietary iron and excessive blood loss being two major environmental contributors. Also, iron deficiency may contain a significant genetic contribution. However, genetic components of iron deficiency are still mainly unexplored.
The goal of the research is to study association between the presence of iron deficiency and haplotypes in selected candidate genes in a multiethnic study population. Candidate genes include genes with reported role in iron metabolism as well as novel genes identified by genome wide association study (GWAS) approach. The proposed work will provide insight into possible genetic contributions to susceptibility or resistance to iron deficiency in humans, and will have important implications for multitude of clinical disorders associated wit perturbations of iron metabolism. Better understanding of variation in iron metabolism and its relationship to clinical manifestations will ultimately enable development of innovative prevention and treatment strategies.
Mouse Studies
Recent advantages in QTL mapping methods in inbred mice allow researchers to make use of existing, dense, publicly available genetic maps to pursue so called “in silico” QTL mapping, and concentrate their resources on phenotyping the animals. Phenotyping multiple animals of the same strain reduce individual variation and results in more precise phenotype measurements.
Previously we measured steady state liver iron content as a phenotype and used publicly available map of 81,032 SNP genotypes in 12 inbred strains for “in silico” QTL mapping. At the same time we used whole genome microarrays to quantify gene expression in liver the same strains to identify differently expressed genes between strains. Genes that correlate with the iron phenotype were identified and compared to the results of “in silico” QTL mapping. The combination of SNP association mapping and gene expression was shown to be able to identify candidate genes underlying liver iron phenotype. We are applying the same approach to identify novel genes related to iron metabolism in duodenum.
Identification of loci involved in iron homeostasis in mice will facilitate understanding of iron homeostasis in humans and will have important implications for numerous clinical disorders associated with disturbances of iron metabolism.